Uploading camera trap records to database
2025-07-23
Source:vignettes/camtrap-upload.Rmd
camtrap-upload.RmdBrief
We need a process that takes (i) camera trap raw data and (ii) site information, and (iii) project information and uploads and processes them to the database in a consistent manner that allows data across projects to be stored together.
Input
There are tree inputs in this process:
- A record table with camera trap records/data (generated from
camtrapR) - A camera information/site information table (generated from field
data, e.g.
proofsafe)
- A single row of details regarding the project (generated manually)
See the appendix for more information on column values in the data uploaded and processed on the database.
Camera trap database model
Once this process is completed, data will be uploaded into the
raw layer of the camtrap schema on the
database. Automatic database views will process this raw data into the
curated layer and processed layer. Zoom in to
the figure below to see details about the data model:
Camera trap database model with the colour signifying the layer of
processing. All data is appended to the raw layer and then flows up
automatically through database views. Data tables in raw and curated
levels can be joined by common keys (e.g. ProjectShortName, SiteID,
SubStation, ID). The colours and position represent the different layers
(yellow = raw, blue = curated, and green =
processed).
Process
The uploading of camera trap data will be undertaken in several steps:
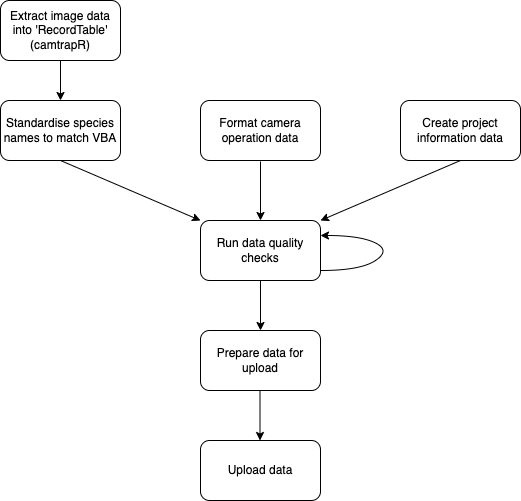
Upload data using the shiny app
As of weda v0.0.1 you can upload data to the database
using an interactive shiny app. You can also view previously uploaded
records on a map and download presence-absences. However, you will still
need to run camtrapR::recordTable() beforehand to
tabularise your image metadata. Thus the section below still might be
relevant.
To run the app from R set up a database connection
(using weda_connect()) and then run the app using
camtrap_app():
Note: to learn more about database connection, see vignette(‘database-connect’)
# Make sure VPN is running
con <- weda::weda_connect(password = keyring::key_get(service = "ari-dev-weda-psql-01",
username = "psql_user"))
weda::camtrap_app(con = con)Upload using R code
Get record table
You should have a directory with camera trap images that have been tagged. All images should be placed in a folder with the title of the station/site. An example of a directory structure for the dummy data is:
#> /home/runner/work/_temp/Library/weda/dummydata/images
#> ├── 2602
#> │ └── 2602__2021-10-17__01-32-47(5).JPG
#> ├── 43134
#> │ └── 43134__2021-11-16__18-15-26(22).JPG
#> ├── 56505
#> │ ├── A
#> │ │ └── 56505__2021-11-30__08-50-17(2).JPG
#> │ └── B
#> │ └── 56505__2021-11-30__08-50-57(59).JPG
#> ├── 832
#> │ └── 832__2021-10-21__14-30-38(8).JPG
#> └── 9941
#> ├── 9941__2021-10-14__18-09-19(13).JPG
#> └── 9941__2021-11-03__14-32-21(11).JPGWe have opted to allow for a two-tiered hierachical system for camera
trap data, with a ‘substation’ folder (e.g. A) allowing to
be nested in a ‘site’ folder (e.g. 56505).
camTrapR denotes this hierachy as Station and
Camera. In our databasing we call the Site and
Substation; with SubStation being nested
within a site. This is useful in cases where you might have multiple
cameras deployed close to one another at a site.
To extract camera trap data we can use the recordTable()
function from camtrapR. The parameters we use for the
function are listed below and if tagging and folder structure is similar
than we can keep it as follows.
Note that depending on the number of metadata tags the number of
columns for this data set may be different. In order for the camera trap
records to meet the standard data format we may need to make some
changes. The metadata tags are denoted with a metadata_
prefix. See recordTableSample for more details.
raw_camtrap_records <- recordTable(inDir = system.file("dummydata/images", package = "weda"),
IDfrom = "metadata",
cameraID = "directory",
stationCol = "SiteID",
camerasIndependent = TRUE,
timeZone = Sys.timezone(location = TRUE),
metadataSpeciesTag = "Species",
removeDuplicateRecords = FALSE,
returnFileNamesMissingTags = TRUE) %>%
rename(SubStation = Camera) %>%
mutate(SubStation = case_when(SiteID == SubStation ~ NA_character_,
TRUE ~ SubStation),
Iteration = 1L)
raw_camtrap_records %>%
kbl() %>%
kable_styling() %>%
scroll_box(width = "100%")| SiteID | SubStation | Species | DateTimeOriginal | Date | Time | delta.time.secs | delta.time.mins | delta.time.hours | delta.time.days | Directory | FileName | metadata_Distance | metadata_Species | n_images | metadata_Individuals | metadata_Behaviour | metadata_Multiples | HierarchicalSubject | Iteration |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2602 | NA | Macropus giganteus | 2021-10-17 01:32:47 | 2021-10-17 | 01:32:47 | 0 | 0 | 0 | 0 | /home/runner/work/_temp/Library/weda/dummydata/images/2602 | 2602__2021-10-17__01-32-47(5).JPG | 2.5 - 5 | Macropus giganteus | 1 | NA | NA | NA | Distance|2.5 - 5, Species|Macropus giganteus | 1 |
| 43134 | NA | Dama dama | 2021-11-16 18:15:26 | 2021-11-16 | 18:15:26 | 0 | 0 | 0 | 0 | /home/runner/work/_temp/Library/weda/dummydata/images/43134 | 43134__2021-11-16__18-15-26(22).JPG | 0 - 2.5 | Dama dama | 1 | Female1 | NA | NA | Distance|0 - 2.5, Individuals|Female1, Species|Dama dama | 1 |
| 56505 | A | Rusa unicolor | 2021-11-30 08:50:17 | 2021-11-30 | 08:50:17 | 0 | 0 | 0 | 0 | /home/runner/work/_temp/Library/weda/dummydata/images/56505/A | 56505__2021-11-30__08-50-17(2).JPG | 2.5 - 5 | Rusa unicolor | 1 | Male1 | NA | NA | Distance|2.5 - 5, Individuals|Male1, Species|Rusa unicolor | 1 |
| 56505 | B | Rusa unicolor | 2021-11-30 08:50:57 | 2021-11-30 | 08:50:57 | 0 | 0 | 0 | 0 | /home/runner/work/_temp/Library/weda/dummydata/images/56505/B | 56505__2021-11-30__08-50-57(59).JPG | 0 - 2.5 | Rusa unicolor | 1 | Male1 | CameraInteraction | NA | Behaviour|CameraInteraction, Distance|0 - 2.5, Individuals|Male1, Species|Rusa unicolor | 1 |
| 832 | NA | Dromaius novaehollandiae | 2021-10-21 14:30:38 | 2021-10-21 | 14:30:38 | 0 | 0 | 0 | 0 | /home/runner/work/_temp/Library/weda/dummydata/images/832 | 832__2021-10-21__14-30-38(8).JPG | 2.5 - 5 | Dromaius novaehollandiae | 1 | NA | NA | NA | Distance|2.5 - 5, Species|Dromaius novaehollandiae | 1 |
| 9941 | NA | Macropus giganteus | 2021-10-14 18:09:19 | 2021-10-14 | 18:09:19 | 0 | 0 | 0 | 0 | /home/runner/work/_temp/Library/weda/dummydata/images/9941 | 9941__2021-10-14__18-09-19(13).JPG | 2.5 - 5 | Macropus giganteus | 1 | NA | NA | 3 | Distance|2.5 - 5, Multiples|3, Species|Macropus giganteus | 1 |
| 9941 | NA | Vulpes vulpes | 2021-11-03 14:32:21 | 2021-11-03 | 14:32:21 | 0 | 0 | 0 | 0 | /home/runner/work/_temp/Library/weda/dummydata/images/9941 | 9941__2021-11-03__14-32-21(11).JPG | 5 - 7.5 | Vulpes vulpes | 1 | NA | NA | NA | Distance|5 - 7.5, Species|Vulpes vulpes | 1 |
Note that with some older models of cameras exifTool and
subsequently camtrapR has difficulty in reading the
date-time format. To fix this you can run the
camtrapR::fixDateTimeOriginal() function on the directory
if needed:
# Optional if using old and problematic cameras:
fixDateTimeOriginal(inDir = system.file("dummydata/images", package = "weda"),
recursive = TRUE)Standardise species names
In our case, species names are Scientific names. In order to
standardise the data we will run a function to append the common names
to the data. The function uses VBA names and the full list of
possibilities can be obtained in the exported dataset:
weda::vba_name_conversions (e.g. run
View(weda::vba_name_conversions)).
Firstly, we can run the function just to check the names for conversion:
standardise_species_names(raw_camtrap_records,
format = "scientific",
speciesCol = "Species",
return_data = FALSE)
#> Warning in standardise_species_names(raw_camtrap_records, format = "scientific", : No match found for Rusa unicolor. Please provide names within the VBA taxa list
#> ✔ Dama dama -> Fallow Deer
#> ✔ Dromaius novaehollandiae -> Emu
#> ✔ Macropus giganteus -> Eastern Grey Kangaroo
#> ✔ Vulpes vulpes -> Red Fox
#> ✖ Rusa unicolorAs seen above, Rusa unicolor is not an accepted scientific name. In the VBA sambar deer are listed as Cervus unicolor. We can change this with code:
raw_camtrap_records_mod <- raw_camtrap_records %>%
mutate(Species = case_when(Species == "Rusa unicolor" ~ "Cervus unicolor",
TRUE ~ Species))
raw_camtrap_records_standardised <- standardise_species_names(raw_camtrap_records_mod,
format = "scientific",
speciesCol = "Species")
#> ✔ Cervus unicolor -> Sambar Deer
#> ✔ Dama dama -> Fallow Deer
#> ✔ Dromaius novaehollandiae -> Emu
#> ✔ Macropus giganteus -> Eastern Grey Kangaroo
#> ✔ Vulpes vulpes -> Red FoxNote that it may still occur that you have issues in converting names
where a common name matches two scientific name. Unfortunately there is
no easy way to fix this, automatically only one name will be chosen but
it may not be the scientific name you wanted. To avoid this then tagging
using scientific names may be worth considering. Otherwise you can
manually edit the output based on the VBA names
weda::vba_name_conversions.
All ticks and no warnings means we can move onto the next step.
Format operation data
Alongside the camera trap records there must be a table that has
details about the camera trap deployment at the site and the location of
the site. This will be site information data users will have obtained
from field sheets or proofsafe. The format of this data is based on the
camera trap operation data used in camtrapR (see
data(camtraps) for an example of this). For our example we
read in an example of this data below with the necessary fields for each
deployment. Alongside reading in we just format columns to correct
classes. Note that the default CRS/epsg for coordinates is
4283.
If you are using Zone 54/55 coordinate reference systems
(28354/28355) the you can use the function
convert_to_latlong(), as long as you have the columns of
Easting, Northing, and Zone (54 or 55) in
your data. It will return your operation table with Latitude
and Longitude.
operationdata <- readr::read_csv(system.file("dummydata/operationdata.csv", package = "weda"),
show_col_types = FALSE) %>%
mutate(SiteID = as.character(SiteID),
Iteration = as.integer(Iteration),
CameraPhotosPerTrigger = as.integer(CameraPhotosPerTrigger))
operationdata %>%
kbl() %>%
kable_styling() %>%
scroll_box(width = "100%")| SiteID | SubStation | Iteration | Latitude | Longitude | DateDeploy | TimeDeploy | DateRetrieve | TimeRetrieve | Problem1_from | Problem1_to | DateTimeDeploy | DateTimeRetrieve | CameraHeight | CameraBearing | CameraSlope | CameraID | CameraModel | CameraSensitivity | CameraDelay | CameraPhotosPerTrigger | CameraQuietPeriod | BaitedUnbaited | BaitType | BaitDistance |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9941 | NA | 1 | -37.24001 | 141.8618 | 2021-10-14 | 14:18:00 | 2021-12-07 | 11:00:00 | NA | NA | 2021-10-14 14:18:00 | 2021-12-07 11:00:00 | 1 | 180 | 2 | HO04101051 | HF2x | Very High | Rapidfire | 5 | 0 | Unbaited | None | NA |
| 2602 | NA | 1 | -36.56451 | 141.1924 | 2021-10-15 | 10:56:00 | 2021-12-07 | 14:51:00 | 2021-11-18 15:56:21 | 2021-12-07 14:51:00 | 2021-10-15 10:56:00 | 2021-12-07 14:51:00 | 1 | 168 | 4 | HO04102687 | HF2x | Very High | Rapidfire | 5 | 0 | Unbaited | None | NA |
| 832 | NA | 1 | -35.85188 | 141.0451 | 2021-10-15 | 15:00:00 | 2021-12-08 | 11:00:00 | NA | NA | 2021-10-15 15:00:00 | 2021-12-08 11:00:00 | 1 | 175 | 2 | HO04102395 | HF2x | Very High | Rapidfire | 5 | 0 | Unbaited | None | NA |
| 43134 | NA | 1 | -36.87991 | 146.0942 | 2021-10-26 | 17:00:00 | 2021-12-21 | 16:21:33 | NA | NA | 2021-10-26 17:00:00 | 2021-12-21 16:21:33 | 1 | 200 | 1 | HO04104906 | HF2x | Very High | Rapidfire | 5 | 0 | Unbaited | None | NA |
| 56505 | A | 1 | -37.27506 | 148.7425 | 2021-10-26 | 10:56:10 | 2021-12-16 | 10:34:51 | NA | NA | 2021-10-26 10:56:10 | 2021-12-16 10:34:51 | 1 | 185 | 8 | HO04104895 | HF2x | Very High | Rapidfire | 5 | 0 | Unbaited | None | NA |
| 56505 | B | 1 | -37.27506 | 148.7425 | 2021-10-26 | 10:56:10 | 2021-12-16 | 10:34:51 | NA | NA | 2021-10-26 10:56:10 | 2021-12-16 10:34:51 | 1 | 180 | 6 | HO04104899 | HF2x | Very High | Rapidfire | 5 | 0 | Unbaited | None | NA |
Create Project Data Row
Alongside the camera data and the camera site/station information we
also want to develop a row with project information that can link to all
cameras and camera trap records for a project. Below we show the columns
that would be in the project dataset. You can check that your project
shortt name isn’t already used by a different project using
check_unique_project(). The data should only be one row and
can easily made with the following code:
projectdata <- tibble(ProjectName = "Dummy Data Project 2023",
ProjectShortName = "DummyData",
DistanceSampling = TRUE,
TerrestrialArboreal = "Terrestrial",
AllSpeciesTagged = TRUE,
DistanceForAllSpecies = TRUE,
ProjectDescription = "Dummy Data for the tutorial on uploading data to the database",
ProjectLeader = "Justin Cally")
projectdata %>%
kbl() %>%
kable_styling() %>%
scroll_box(width = "100%")| ProjectName | ProjectShortName | DistanceSampling | TerrestrialArboreal | AllSpeciesTagged | DistanceForAllSpecies | ProjectDescription | ProjectLeader |
|---|---|---|---|---|---|---|---|
| Dummy Data Project 2023 | DummyData | TRUE | Terrestrial | TRUE | TRUE | Dummy Data for the tutorial on uploading data to the database | Justin Cally |
Data checks
At this stage it is important to look at our three datasets and run
data quality checks on them to ensure that they have sufficient data and
are able to be properly linked. For this we have developed a single R
function to fun the checks (camera_trap_dq()). This
function uses pointblank R package to run extensive data
checks. For data to be of sufficient quality it must pass all checks.
The output of camera_trap_dq() provides three data quality
statements (one for each table). You can use this to help identify
errors in the data to fix before preparing the data for upload.
dq <- camera_trap_dq(camtrap_records = raw_camtrap_records_standardised,
camtrap_operation = operationdata,
project_information = projectdata)
#> Automatically standardising column classes, see weda::data_dictionary for database column classes
#> Warning: There was 1 warning in `dplyr::mutate()`.
#> ℹ In argument: `dplyr::across(...)`.
#> Caused by warning:
#> ! NAs introduced by coercion
dq[[1]]
dq[[2]]| Pointblank Validation | |||||||||||||
| [2025-07-23|05:50:59]
tibble
camtrap_operationWARN
—
STOP
1
NOTIFY
—
|
|||||||||||||
| STEP | COLUMNS | VALUES | TBL | EVAL | UNITS | PASS | FAIL | W | S | N | EXT | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 2 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 3 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 4 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 5 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 6 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 7 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 8 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 9 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 10 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 11 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 12 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 13 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 14 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 15 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 16 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 17 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 18 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 19 | col_exists()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 20 | rows_distinct()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 21 | col_is_character()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 22 | col_is_character()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 23 | col_is_character()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 24 | col_is_character()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 25 | col_is_character()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 26 | col_is_character()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 27 | col_is_numeric()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 28 | col_is_numeric()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 29 | col_is_numeric()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 30 | col_is_date()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 31 | col_is_date()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 32 | col_is_integer()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 33 | col_is_integer()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 34 | col_is_posix()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 35 | col_is_posix()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 36 | col_is_posix()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 37 | col_is_posix()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 38 | col_vals_in_set()
|
✓ | 6 |
61.00
|
00.00
|
○ | ○ | — | — | ||||
| 39 | col_vals_in_set()
|
✓ | 6 |
61.00
|
00.00
|
○ | ○ | — | — | ||||
| 40 | col_vals_between()
|
✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | ||||
| 41 | col_vals_between()
|
✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | ||||
| 42 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 43 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 44 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 45 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 46 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 47 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 48 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 49 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 50 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 51 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 52 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 53 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 54 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 55 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 56 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 57 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 58 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 59 | col_vals_not_null()
|
— | ✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | |||
| 60 | col_vals_in_set()
|
✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | ||||
| 61 | col_vals_in_set()
|
✓ | 6 |
61.00
|
00.00
|
— | ○ | — | — | ||||
| 2025-07-23 05:50:59 UTC 1.3 s 2025-07-23 05:51:00 UTC | |||||||||||||
dq[[3]]| Pointblank Validation | |||||||||||||
| [2025-07-23|05:51:00]
tibble
project_informationWARN
—
STOP
1
NOTIFY
—
|
|||||||||||||
| STEP | COLUMNS | VALUES | TBL | EVAL | UNITS | PASS | FAIL | W | S | N | EXT | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | row_count_match()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 2 | col_vals_not_null()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 3 | col_vals_not_null()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 4 | col_vals_not_null()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 5 | col_vals_not_null()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 6 | col_vals_not_null()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 7 | col_vals_not_null()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 8 | col_vals_not_null()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 9 | col_vals_not_null()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 10 | col_is_logical()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 11 | col_is_logical()
|
— | ✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | |||
| 12 | col_vals_in_set()
|
✓ | 1 |
11.00
|
00.00
|
— | ○ | — | — | ||||
| 2025-07-23 05:51:00 UTC < 1 s 2025-07-23 05:51:01 UTC | |||||||||||||
As you can see from the above data quality checks there is an issue we should fix before re-running the data quality:
- In the camera trap records
metadata_Multipleshas some missing values.
For the second point when there was only one individual in a photo it was not tagged with an integer. In these cases it should be 1. To fix these issues we could run:
raw_camtrap_records_fixed <- raw_camtrap_records_standardised %>%
mutate(metadata_Multiples = coalesce(as.integer(metadata_Multiples), 1L))
#> Warning: There was 1 warning in `mutate()`.
#> ℹ In argument: `metadata_Multiples = coalesce(as.integer(metadata_Multiples),
#> 1L)`.
#> Caused by warning in `list2()`:
#> ! NAs introduced by coercion
dq2 <- camera_trap_dq(camtrap_records = raw_camtrap_records_fixed,
camtrap_operation = operationdata,
project_information = projectdata)
#> Automatically standardising column classes, see weda::data_dictionary for database column classes
# We can check it is passing all tests with:
all(sapply(dq2, function(x) all(x[["validation_set"]][["all_passed"]])))
#> [1] TRUEPrepare data for upload
Once all data quality issues are fixed, and only when all data
quality issues are fixed you can prepare your data for upload to the
database. This process (prepare_camtrap_upload()) will
generate IDs for your records (to avoid duplicate records on the
database) and properly format the data:
data_for_upload <- prepare_camtrap_upload(dq2)Upload the data
Note that before uploading data you will need to have an
established connection to the database (e.g. con) in your
R environment. See the vignette
on database connection for more details
Using the data prepared for upload made above
(data_for_upload) you can upload the the data in one
line:
con <- weda_connect(password = keyring::key_get(service = "ari-dev-weda-psql-01",
username = "psql_user"))
upload_camtrap_data(con = con,
data_list = data_for_upload,
uploadername = "Justin Cally")The upload may take several minutes as it creates
materialized views, which require some processing. Be
patient and leave your R session running.
Automatic Database Views
Once uploaded, the postgresql database will append your
data to several easy to interact with views:
Curated Layer
The curated layer is basic processing with duplicates removed and most/all data fields retained:
-
curated_camtrap_operation: Is a table that stores the most recent entries for all camera trap deployments across projects (i.e. no duplicate sites)
-
curated_camtrap_records: Is a table that stores the most recent camera trap records across projects (i.e. no duplicate images). This is based on a unique image being determined based on the project, site, substation, date-time and filename.
-
curated_project_information: Is a table that stores the most recent project information entries (i.e. no duplicate projects)
Processed Layer
The processed layer is more heavily processed with duplicates removed and joins, summaries and more succinct tables:
processed_site_substation_presence_absence: Is a table with the presence and absence of each species at each site. The possible absent species from each site are only derived from the species pool for a given project. This avoids absences of species for particular projects that did not set out to record that species.processed_site_substation_daily_presence_absence: As above but now all presences and absences are daily. Includes early truncation of camera deployment from thecurated_camtrap_operationdataset when there is a problem with the operation period.
# This is how you write the views. It only needs to be done once:
DBI::dbExecute(conn = con, paste(DBI::SQL("CREATE VIEW camtrap.curated_camtrap_records AS"), records_curated_view(con)))
DBI::dbExecute(conn = con, paste(DBI::SQL("CREATE VIEW camtrap.curated_camtrap_operation AS"), operation_curated_view(con)))
DBI::dbExecute(conn = con, paste(DBI::SQL("CREATE VIEW camtrap.curated_project_information AS"), project_curated_view(con)))
DBI::dbExecute(conn = con, paste(DBI::SQL("CREATE VIEW camtrap.processed_site_substation_presence_absence AS"), processed_SubStation_presence_absence(con = con, return_data = FALSE)))
DBI::dbExecute(conn = con, paste(DBI::SQL("CREATE VIEW camtrap.processed_site_substation_daily_presence_absence AS"), processed_SubStation_presence_absence(con = con, daily = TRUE, return_data = FALSE)))Appendix
A data dictionary is provided in this package
(data(data_dictionary)) and also available in the
data_dictionary schema. Below is the data dictionary for
the camtrap schema:
data_dictionary %>%
filter(schema == "camtrap") %>%
select(table_name, table_description, column_name, column_class, column_description) %>%
kbl() %>%
kable_styling(c("condensed"), full_width = F) %>%
collapse_rows(1:3, valign = "top") %>%
scroll_box(width = "100%", height = "1000px")| table_name | table_description | column_name | column_class | column_description |
|---|---|---|---|---|
| vba_name_conversions | NA | taxon_id | numeric | NA |
| scientific_name | character | Scientific taxa name as per VBA | ||
| common_name | character | Common taxa name as per VBA | ||
| raw_camtrap_operation | Is a table that stores ALL camera trap deployments across projects (i.e. duplicates allowes). This is the table where uploads are appended to. | ProjectShortName | character | Short project name that data was collected for. Approximately five words in lowercase seperated by underscores. |
| SiteID | character | Site identification. Can be camera trap location or have a nesting of substations. Project specific. | ||
| SubStation | character | Nested location of camera trap within a site. Used in cases where a site has multiple camera trap deployments. | ||
| geohash | character | 8-digit geohash code based on longitude and latitude | ||
| Iteration | integer | Useful for multi-season surveys, this is the nth deployment iteration. If it is the first survey at a site, use 1. | ||
| Latitude | numeric | Latitude of camera in decimal degrees (EPSG: 4283) | ||
| Longitude | numeric | Longitude of camera in decimal degrees (EPSG: 4283) | ||
| DateDeploy | Date | Date camera was deployed | ||
| TimeDeploy | character | Time camera was deployed | ||
| DateRetrieve | Date | Date camera was retrieved | ||
| TimeRetrieve | character | Time camera was retrieved | ||
| DateTimeDeploy | POSIXct, POSIXt | Date-time camera was deployed | ||
| DateTimeRetrieve | POSIXct, POSIXt | Date-time camera was deployed | ||
| Problem1_from | POSIXct, POSIXt | If there was a problem with the camera, when (date-time) did it start | ||
| Problem1_to | POSIXct, POSIXt | If there was a problem with the camera, when (date-time) did it end (usually when camera is picked up) | ||
| CameraHeight | numeric | Height above ground of camera (in metres) | ||
| CameraBearing | numeric | The bearing (in degrees) of the camera (0-360) | ||
| CameraSlope | numeric | The slope of terrain the camera is on (in percentage) | ||
| CameraID | character | ID of camera (optional) | ||
| CameraSensitivity | character | Sensitivity of camera (low, medium, high or very high) | ||
| CameraDelay | character | Delay of camera in taking photos (Rapidfire or time in seconds) | ||
| CameraPhotosPerTrigger | integer | Number of photos per trigger (e.g. 5) | ||
| CameraQuietPeriod | numeric | The time in between triggers to wait before taking more photos. Usually ‘No Delay’, which is listed here as 0 | ||
| BaitedUnbaited | character | Whether camera was Baited or Unbaited | ||
| BaitType | character | Type of bait used: None, Creamed Honey, Small Mammal Bait, Predator Bait (i.e, meat bait), Non-toxic curiosity bait, Toxic curiosity bait, Predator Lure (i.e., urine, faeces, etc.) or Other | ||
| BaitDistance | numeric | Distance of the camera to the bait (in metres) | ||
| CameraModel | character | Model of camera | ||
| camtrap_operation_database_ID | character | Unique ID of the cameratrap record. ID is formulated from key variables of ProjectShortName, SiteID, SubStation | ||
| Timestamp | POSIXct, POSIXt | Time/date of upload (should exist as Greenwhich Mean Time) | ||
| Uploader | character | Name of person uploading data | ||
| raw_camtrap_records | Is a table that stores ALL camera trap records across projects (i.e. duplicate images allowed). This is the table where uploads are appended to. | ProjectShortName | character | Short project name that data was collected for. Approximately five words in lowercase seperated by underscores. |
| SiteID | character | Site identification. Can be camera trap location or have a nesting of substations. Project specific. | ||
| SubStation | character | Nested location of camera trap within a site. Used in cases where a site has multiple camera trap deployments. | ||
| Iteration | integer | Useful for multi-season surveys, this is the nth deployment iteration. If it is the first survey at a site, use 1. | ||
| scientific_name | character | Scientific taxa name as per VBA | ||
| DateTimeOriginal | POSIXct, POSIXt | Date and time of photo-capture | ||
| Date | Date | Date of photo | ||
| Time | character | Time of photo | ||
| delta.time.secs | numeric | Time lag between images (seconds) | ||
| delta.time.mins | numeric | Time lag between images (minutes) | ||
| delta.time.hours | numeric | Time lag between images (hours) | ||
| delta.time.days | numeric | Time lag between images (days) | ||
| Directory | character | Local directory path image was stored when metadata extracted | ||
| FileName | character | Filename of image when metadata extracted | ||
| metadata_Distance | character | Optional metadata field for distance bin (in metres) | ||
| metadata_Species | character | Metadata field for what species was used when tagging | ||
| metadata_Multiples | integer | Mandatory metadata field for number of individuals in the photo | ||
| n_images | integer | Numer of images for record | ||
| metadata_Individuals | character | Metadata field for sex/age classification (e.g. Male1, Male2 for two males) | ||
| HierarchicalSubject | character | Full metadata string from the photo extracted by camtrapR | ||
| metadata_Behaviour | character | Optional metadata field for important behaviour (e.g. marker interaction) | ||
| common_name | character | Common taxa name as per VBA | ||
| camtrap_record_database_ID | character | Unique ID of the cameratrap record. ID is formulated from key variables of ProjectShortName, SiteID, SubStation, DateTimeOriginal, FileName | ||
| Timestamp | POSIXct, POSIXt | Time/date of upload (should exist as Greenwhich Mean Time) | ||
| Uploader | character | Name of person uploading data | ||
| raw_project_information | Is a table that stores ALL project information entries (i.e. duplicate projects allowed). This is the table where uploads are appended to. | ProjectName | character | Longer project name (as per official documents) |
| ProjectShortName | character | Short project name that data was collected for. Approximately five words in lowercase seperated by underscores. | ||
| DistanceSampling | logical | Logical flag (TRUE/FALSE), whether distance sampling was done for project | ||
| TerrestrialArboreal | character | Whether camera was Terrestrial or Arboreal | ||
| AllSpeciesTagged | logical | Logical flag (TRUE/FALSE), whether all species seen were tagged | ||
| DistanceForAllSpecies | logical | Whether or not distance has been tagged for all species recorded | ||
| ProjectDescription | character | Short description of the project | ||
| ProjectLeader | character | Name of the person/persons responsible for the project | ||
| camtrap_project_database_ID | character | Unique ID of the camera trap project ID is formulated from key variables of ProjectShortName | ||
| Timestamp | POSIXct, POSIXt | Time/date of upload (should exist as Greenwhich Mean Time) | ||
| Uploader | character | Name of person uploading data | ||
| spatial_camtrap_operation | NA | camtrap_operation_database_ID | character | Unique ID of the cameratrap record. ID is formulated from key variables of ProjectShortName, SiteID, SubStation |
| ProjectShortName | character | Short project name that data was collected for. Approximately five words in lowercase seperated by underscores. | ||
| SiteID | character | Site identification. Can be camera trap location or have a nesting of substations. Project specific. | ||
| SubStation | character | Nested location of camera trap within a site. Used in cases where a site has multiple camera trap deployments. | ||
| geohash | character | 8-digit geohash code based on longitude and latitude | ||
| Iteration | integer | Useful for multi-season surveys, this is the nth deployment iteration. If it is the first survey at a site, use 1. | ||
| Latitude | numeric | Latitude of camera in decimal degrees (EPSG: 4283) | ||
| Longitude | numeric | Longitude of camera in decimal degrees (EPSG: 4283) | ||
| DateDeploy | Date | Date camera was deployed | ||
| TimeDeploy | character | Time camera was deployed | ||
| DateRetrieve | Date | Date camera was retrieved | ||
| TimeRetrieve | character | Time camera was retrieved | ||
| DateTimeDeploy | POSIXct, POSIXt | Date-time camera was deployed | ||
| DateTimeRetrieve | POSIXct, POSIXt | Date-time camera was deployed | ||
| Problem1_from | POSIXct, POSIXt | If there was a problem with the camera, when (date-time) did it start | ||
| Problem1_to | POSIXct, POSIXt | If there was a problem with the camera, when (date-time) did it end (usually when camera is picked up) | ||
| CameraHeight | numeric | Height above ground of camera (in metres) | ||
| CameraBearing | numeric | The bearing (in degrees) of the camera (0-360) | ||
| CameraSlope | numeric | The slope of terrain the camera is on (in percentage) | ||
| CameraID | character | ID of camera (optional) | ||
| CameraSensitivity | character | Sensitivity of camera (low, medium, high or very high) | ||
| CameraDelay | character | Delay of camera in taking photos (Rapidfire or time in seconds) | ||
| CameraPhotosPerTrigger | integer | Number of photos per trigger (e.g. 5) | ||
| CameraQuietPeriod | numeric | The time in between triggers to wait before taking more photos. Usually ‘No Delay’, which is listed here as 0 | ||
| BaitedUnbaited | character | Whether camera was Baited or Unbaited | ||
| BaitType | character | Type of bait used: None, Creamed Honey, Small Mammal Bait, Predator Bait (i.e, meat bait), Non-toxic curiosity bait, Toxic curiosity bait, Predator Lure (i.e., urine, faeces, etc.) or Other | ||
| BaitDistance | numeric | Distance of the camera to the bait (in metres) | ||
| CameraModel | character | Model of camera | ||
| Timestamp | POSIXct, POSIXt | Time/date of upload (should exist as Greenwhich Mean Time) | ||
| Uploader | character | Name of person uploading data | ||
| geometry | pq_geometry | NA |